I am a PhD student at Texas A&M University, advised by Dr. Nancy M. Amato. My research centers on motion planning algorithms applied to robotics and computational biology. I have studied how workspace guidance can be leveraged to improve runtime efficiency and space coverage of motion planning algorithms in robotics and computational biology applications. I am passionate about education and broadening participation in computing.
Special Thank you!
The University of Illinois at Urbana-Champaign where I have been a visiting PhD student since Summer 2019. The Computer Science Department has been a great research community for me.Personal Update
Google Scholar
Research Statement
Teaching Statement
Diversity Statement
Publications
Current Mentees
Undergraduates: Ana Elissa Cabrera, Melissa Aninagyei-Bonsu, Vasu Chalassani, Keerthana Nallamatou, Matthew Rizen
Graduates: Eunice Chan Ananya Yammanuru
Publications
Hierarchical Annotated Skeleton-Guided Tree-based Motion Planning, Diane Uwacu, Ananya Yammanuru, Keerthana Nallamotu, Vasu Chalasani, Marco Morales, Nancy M. Amato, arXiv Preprint, Sep 2023. @misc{uwacu2023hierarchical, We present a hierarchical tree-based motion planning strategy, HAS-RRT, guided by the workspace skeleton to solve motion planning problems in robotics and computational biology. Relying on the information about the connectivity of the workspace and the ranking of available paths in the workspace, the strategy prioritizes paths indicated by the workspace guidance to find a valid motion plan for the moving object efficiently. In instances of suboptimal guidance, the strategy adapts its reliance on the guidance by hierarchically reverting to local exploration of the planning space. We offer an extensive comparative analysis against other tree-based planning strategies and demonstrate that HAS-RRT reliably and efficiently finds low-cost paths. In contrast to methods prone to inconsistent performance across different environments or reliance on specific parameters, HAS-RRT is robust to workspace variability.
Keywords: Lazy Planning, Motion Planning, Workspace Topology
Links : [ArXiv] BibTex
title={Hierarchical Annotated Skeleton-Guided Tree-based Motion Planning},
author={Diane Uwacu and Ananya Yammanuru and Keerthana Nallamotu and Vasu Chalasani and Marco Morales and Nancy M. Amato},
year={2023},
eprint={2309.10801},
archivePrefix={arXiv},
primaryClass={cs.RO}
}Abstract
Scalable Multi-robot Motion Planning for Congested Environments With Topological Guidance, Courtney McBeth, James Motes, Diane Uwacu, Marco Morales, Nancy M. Amato, In IEEE Robotics and Automation Letters, Aug 2023. DOI: 10.1109/LRA.2023.3312980 @ARTICLE{10243143, Multi-robot motion planning (MRMP) is the problem of finding collision-free paths for a set of robots in a continuous state space. The difficulty of MRMP increases with the number of robots and is exacerbated in environments with narrow passages that robots must pass through, like warehouse aisles where coordination between robots is required. In single-robot settings, topology-guided motion planning methods have shown improved performance in these constricted environments. In this work, we extend an existing topology-guided single-robot motion planning method to the multi-robot domain to leverage the improved efficiency provided by topological guidance. We demonstrate our method's ability to efficiently plan paths in complex environments with many narrow passages, scaling to robot teams of size up to 25 times larger than existing methods in this class of problems. By leveraging knowledge of the topology of the environment, we also find higher-quality solutions than other methods.
Keywords: Motion Planning, Multi-Agent, Workspace Topology
Links : [Published] [ArXiv] [Video] BibTex
author={McBeth, Courtney and Motes, James and Uwacu, Diane and Morales, Marco and Amato, Nancy M.},
journal={IEEE Robotics and Automation Letters},
title={Scalable Multi-robot Motion Planning for Congested Environments With Topological Guidance},
year={2023},
volume={},
number={},
pages={1-8},
doi={10.1109/LRA.2023.3312980}}Abstract
Hierarchical Planning With Annotated Skeleton Guidance, Diane Uwacu, Ananya Yammanuru, Marco Morales, Nancy M. Amato, IEEE Robotics and Automation Letters (RA-L), Vol: 7, Issue: 4, pp. 11055-11061, Oct 2022. DOI: 10.1109/LRA.2022.3196885 @ARTICLE{9851528, We present a hierarchical skeleton-guided motion planning algorithm to guide mobile robots. A good skeleton maps the connectivity of the subspace of c-space containing significant degrees of freedom and is able to guide the planner to find the desired solutions fast. However, sometimes the skeleton does not closely represent the free c-space, which often misleads current skeleton-guided planners. The hierarchical skeleton-guided planning strategy gradually relaxes its reliance on the workspace skeleton as C-space is sampled, thereby incrementally returning a sub-optimal path, a feature that is not guaranteed in the standard skeleton-guided algorithm. Experimental comparisons to the standard skeleton guided planners and other lazy planning strategies show significant improvement in roadmap construction run time while maintaining path quality for multi-query problems in cluttered environments.
Keywords: Lazy Evaluation, Motion Planning, Workspace Topology
Links : [Published] [Manuscript] [Video] BibTex
author={Uwacu, Diane and Yammanuru, Ananya and Morales, Marco and Amato, Nancy M.},
journal={IEEE Robotics and Automation Letters},
title={Hierarchical Planning With Annotated Skeleton Guidance},
year={2022},
volume={7},
number={4},
pages={11055-11061},
doi={10.1109/LRA.2022.3196885}}Abstract
Evaluating Guiding Spaces for Motion Planning, Amnon Attali, Stav Ashur, Isaac Burton Love, Courtney McBeth, James Motes, Diane Uwacu, Marco Morales, Nancy M. Amato, IROS 2022, Workshop for Evaluating Motion Planning Performance, Kyoto, Japan, Oct 2022. @misc{https://doi.org/10.48550/arxiv.2210.08640, Randomized sampling based algorithms are widely used in robot motion planning due to the problem's intractability, and are experimentally effective on a wide range of problem instances. Most variants do not sample uniformly at random, and instead bias their sampling using various heuristics for determining which samples will provide more information, or are more likely to participate in the final solution. In this work, we define the motion planning guiding space, which encapsulates many seemingly distinct prior works under the same framework. In addition, we suggest an information theoretic method to evaluate guided planning which places the focus on the quality of the resulting biased sampling. Finally, we analyze several motion planning algorithms in order to demonstrate the applicability of our definition and its evaluation.
Keywords: Algorithms, Guidance
Links : [ArXiv] BibTex
doi = {10.48550/ARXIV.2210.08640},
url = {https://arxiv.org/abs/2210.08640},
author = {Attali, Amnon and Ashur, Stav and Love, Isaac Burton and McBeth, Courtney and Motes, James and Uwacu, Diane and Morales, Marco and Amato, Nancy M.},
keywords = {Robotics (cs.RO), Artificial Intelligence (cs.AI), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {Evaluating Guiding Spaces for Motion Planning},
publisher = {arXiv},
year = {2022},
copyright = {Creative Commons Attribution 4.0 International}
}
Abstract
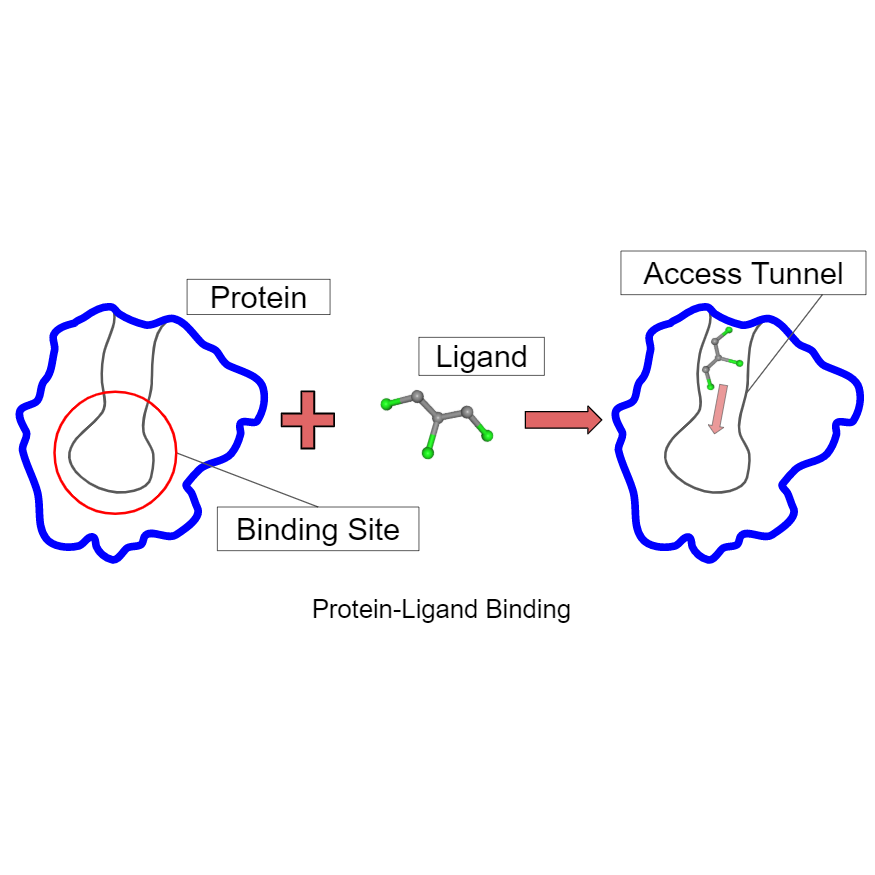
Topology-Guided Roadmap Construction with Dynamic Region Sampling, Read Sandström, Diane Uwacu, Jory Denny, Nancy M. Amato, IEEE Robotics and Automation Letters (RA-L), vol. 5, no. 4,, pp. 6161-6168, virtual, Oct 2020. DOI: 10.1109/LRA.2020.3010487 @ARTICLE{9144398, Many types of planning problems require discovery of multiple pathways through the environment, such as multi-robot coordination or protein ligand binding. The Probabilistic Roadmap (PRM) algorithm is a powerful tool for this case, but often cannot efficiently connect the roadmap in the presence of narrow passages. In this letter, we present a guidance mechanism that encourages the rapid construction of well-connected roadmaps with PRM methods. We leverage a topological skeleton of the workspace to track the algorithm's progress in both covering and connecting distinct neighborhoods, and employ this information to focus computation on the uncovered and unconnected regions. We demonstrate how this guidance improves PRM's efficiency in building a roadmap that can answer multiple queries in both robotics and protein ligand binding applications.
Keywords: Sampling-Based Motion Planning, Workspace Topology
Links : [Published] BibTex
author={R. {Sandström} and D. {Uwacu} and J. {Denny} and N. M. {Amato}},
journal={IEEE Robotics and Automation Letters},
title={Topology-Guided Roadmap Construction With Dynamic Region Sampling},
year={2020},
volume={5},
number={4},
pages={6161-6168},}Abstract
Using Guided Motion Planning to Study Binding Site Accessibility, Diane Uwacu, Abigail Ren, Shawna Thomas, Nancy M. Amato, Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Issue: 109, (Virtual) New York, USA, Sep 2020. DOI: 10.1145/3388440.3414707 @inbook{10.1145/3388440.3414707, Computational methods are commonly used to predict protein-ligand interactions. These methods typically search for regions with favorable energy that geometrically fit the ligand, and then rank them as potential binding sites. While this general strategy can provide good predictions in some cases, it does not do well when the binding site is not accessible to the ligand. In addition, recent research has shown that in some cases protein access tunnels play a major role in the activity and stability of the protein's binding interactions. Hence, to fully understand the binding behavior of such proteins, it is imperative to identify and study their access tunnels. In this work, we present a motion planning algorithm that scores protein binding site accessibility for a particular ligand. This method can be used to screen ligand candidates for a protein by eliminating those that cannot access the binding site. This method was tested on two case studies to analyze effects of modifying a protein's access tunnels to increase activity and/or stability as well as study how a ligand inhibitor blocks access to the protein binding site.
Keywords: Computational Biology, Ligand Binding, Motion Planning
Links : [Published] [Manuscript] BibTex
author = {Uwacu, Diane and Ren, Abigail and Thomas, Shawna and Amato, Nancy M.},
title = {Using Guided Motion Planning to Study Binding Site Accessibility},
year = {2020},
isbn = {9781450379649},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3388440.3414707},
abstract = {Computational methods are commonly used to predict protein-ligand interactions. These methods typically search for regions with favorable energy that geometrically fit the ligand, and then rank them as potential binding sites. While this general strategy can provide good predictions in some cases, it does not do well when the binding site is not accessible to the ligand. In addition, recent research has shown that in some cases protein access tunnels play a major role in the activity and stability of the protein's binding interactions. Hence, to fully understand the binding behavior of such proteins, it is imperative to identify and study their access tunnels. In this work, we present a motion planning algorithm that scores protein binding site accessibility for a particular ligand. This method can be used to screen ligand candidates for a protein by eliminating those that cannot access the binding site. This method was tested on two case studies to analyze effects of modifying a protein's access tunnels to increase activity and/or stability as well as study how a ligand inhibitor blocks access to the protein binding site.},
booktitle = {Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics},
articleno = {109},
numpages = {10}
}
Abstract
Using Motion Planning to Evaluate Protein Binding Site Accessibility, Diane Uwacu, Everett Yang, Shawna Thomas, Nancy M. Amato, Technical Report, TR18-001, Parasol Laboratory, Department of Computer Science, Texas A&M University, College Station TX 77848, USA, Jul 2018. N/A Despite many efforts and considerable breakthroughs in ligand binding prediction, the best predictors still produce many false positives and a reliable,
fully automated prediction framework has yet to be developed. Binding site accessibility is an important feature ignored by methods that classify binding based solely on the energetic or geometric properties of the final bound protein-ligand complex. To evaluate this necessity, we transform the ligand accessibility problem into a robot motion planning problem where the ligand is modeled as a flexible agent whose task is to travel from outside the protein to its binding site. We use Rapidly-exploring Random Graphs coupled with Mean Curve workspace skeletons to quickly and thoroughly explore a protein environment in order to produce valid paths for ligand motion. Path weights reflect the influences of intermolecular forces on the given ligand. Low weight paths are extracted and analyzed for characteristics of accessibility. In this paper, we show that our algorithm provides a mechanism to evaluate binding site accessibility for a ligand.
Keywords: Ligand Binding, Motion Planning, Workspace Topology
Links : [Manuscript] BibTex
Abstract
The Impact of Approximate Methods on Local Learning in Motion Planning, Diane Uwacu, Chinwe Ekenna, Shawna Thomas, Nancy Amato, 1st International Workshop on Robot Learning and Planning (RLP 2016), in conjunction with RSS 2016, pp. 38-44, Ann Arbor, MI, Jun 2016. @inproceedings{uwacu-wrlp-2016 Machine learning methods have been applied to many motion planning algorithms including probabilistic roadmap methods (PRM). There are many variants of these methods and choosing the best one every time is hard and depends on local properties of the environment. A successful learning approach has been developed to offset this issue. This learning approach was applied to PRMs to help decide intelligently what method to utilize in dynamically created local regions of the environment or task space. It used exact neighbor finding approaches and removed the need to partition environments to get improved results. In this work we make further advances by introducing approximate neighbor finder methods. It has been established that approximate neighbor finding methods are faster than exact methods, still work well in connecting nodes to edges in PRMs, and that connection is robust to noise. We study what happens when noise is introduced into learning by using approximate methods instead of already studied exact methods. We show that the impact of noise on learning depends on how much learning needs to take place given the topology of the environment. Our results demonstrate a correlation between heterogeneity and the need for learning over a local region.
Keywords: Machine Learning, Sampling-Based Motion Planning
Links : [ArXiv] [Manuscript] BibTex
, author = "Diane Uwacu and Chinwe Ekenna and Shawna Thomas and Nancy M. Amato"
, title = "The Impact of Approximate Methods on Local Learning in Motion Planning"
, booktitle = "1st International Workshop on Robot Learning and Planning (RLP 2016), in conjunction with RSS 2016"
, month = "June"
, year = "2016"
, note = "http://chitsazlab.org/robotics/rlp2016/RLP_2016_paper_11.html"
}
Abstract
Improved Roadmap Connection via Local Learning for Sampling Based Planners, Chinwe Ekenna, Diane Uwacu, Shawna Thomas, Nancy Amato, Proc. IEEE/RSJ Int. Conf. Intel. Rob. Syst. (IROS), pp. 3227-3234, Hamburg, Germany, Oct 2015. DOI: 10.1109/IROS.2015.7353825 @INPROCEEDINGS{7353825, author={C. {Ekenna} and D. {Uwacu} and S. {Thomas} and N. M. {Amato}}, booktitle={2015 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS)}, title={Improved roadmap connection via local learning for sampling based planners}, year={2015}, volume={}, number={}, pages={3227-3234},} Probabilistic Roadmap Methods (PRMs) solve the motion planing problem by constructing a roadmap (or graph) that models the motion space when feasible local motions exist. PRMs and variants contain several phases during roadmap generation i.e., sampling, connection, and query. Some work has been done to apply machine learning to the connection phase to decide which variant to employ, but it uses a global learning approach that is inefficient in heterogeneous situations. We present an algorithm that instead uses local learning: it only considers the performance history in the vicinity of the current connection attempt and uses this information to select good candidates for connection. It thus removes any need to explicitly partition the environment which is burdensome and typically difficult to do. Our results show that our method learns and adapts in heterogeneous environments, including a KUKA youBot with a fixed and mobile base. It finds solution paths faster for single and multi-query scenarios and builds roadmaps with better coverage and connectivity given a fixed amount of time in a wide variety of input problems. In all cases, our method outperforms the previous adaptive connection method and is comparable or better than the best individual method.
Keywords: Machine Learning, Motion Planning, Sampling-Based Motion Planning
Links : [Published] BibTex
Abstract
Studying Learning Techniques in Different Phases of PRM Construction, Chinwe Ekenna, Diane Uwacu, Shawna Thomas, Nancy Amato, In Machine Learning in Planning and Control of Robot Motion Workshop (IROS-MLPC), Hamburg, Germany, Oct 2015. DOI: 10.1109/IROS.2015.7353825 no bib for now. Probabilistic Roadmap Methods (PRMs) solve the motion planning problem in two phases by sampling free configurations and connecting them together to build a map that is used to find a valid path. Existing algorithms are highly sensitive to the topology of the problem, and their efficiency depends on applying them to a compatible problem. Reinforcement learning has been applied to motion planning and rewards the action performed by planners during either sampling or connection, but not both. Previous work computed a global reward and action scheme, which saw a setback when heterogeneous environments were concerned. Local learning (connection) was recently introduced to offset this weakness identified during global learning, and there was some improvement in planner performance. These different learning schemes (global and local) have shown strengths and weaknesses individually. In this paper, we investigate local learning for sampling. We study what type of learning to apply when, and how the two phases of PRM roadmap construction interact, which has not been investigated before. We show the performance using each scheme on a KUKAyouBot, an 8 degree of freedom robot, and analyze what happens when they are all combined during roadmap construction
Keywords: Machine Learning, Sampling-Based Motion Planning
Links : [Published] BibTex
Abstract