
Prediction of protein-ligand interaction, or binding, is important for drug discovery research. Specifially, the research aims to answer two questions: (1) Can the ligand access the binding site of the target protein? (2) Is the protein-ligand interaction stable?
We use motion planningn algorithms to study binding site accessibility. The protein is modeled as an environment and the ligand as the moving object or robot with a goal to reach the binding site.
Using Topological Guidance to Study Binding Site Accessibility
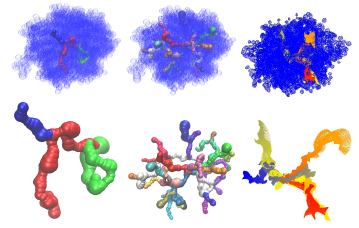
We use Rapidly-exploring Random Graphs coupled with Mean Curve workspace skeletons to quickly and thoroughly explore a protein environment and find valid paths for ligand motion. We annotated the mean curvature skeleton of the protein with energy information, and then bias planning to explore regions with favorabke energy values first. We used our algorithm to analyze accessibility of the protein Haloalkane dehalogenase (dhaA) from the bacterium Rhodococcus rhodochrous, used in soil inoculation. The protein has multiple mutants whose binding activity is regulated by the accessibility of the buried binding site.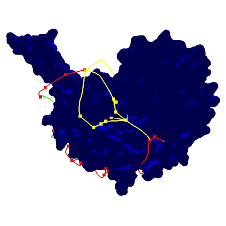
Related Publications
Using Guided Motion Planning to Study Binding Site Accessibility, Diane Uwacu, Abigail Ren, Shawna Thomas, Nancy M. Amato, Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Issue: 109, (Virtual) New York, USA, Sep 2020. DOI: 10.1145/3388440.3414707 @inbook{10.1145/3388440.3414707, Computational methods are commonly used to predict protein-ligand interactions. These methods typically search for regions with favorable energy that geometrically fit the ligand, and then rank them as potential binding sites. While this general strategy can provide good predictions in some cases, it does not do well when the binding site is not accessible to the ligand. In addition, recent research has shown that in some cases protein access tunnels play a major role in the activity and stability of the protein's binding interactions. Hence, to fully understand the binding behavior of such proteins, it is imperative to identify and study their access tunnels. In this work, we present a motion planning algorithm that scores protein binding site accessibility for a particular ligand. This method can be used to screen ligand candidates for a protein by eliminating those that cannot access the binding site. This method was tested on two case studies to analyze effects of modifying a protein's access tunnels to increase activity and/or stability as well as study how a ligand inhibitor blocks access to the protein binding site.
Keywords: Computational Biology, Ligand Binding, Motion Planning
Links : [Published] [Manuscript] BibTex
author = {Uwacu, Diane and Ren, Abigail and Thomas, Shawna and Amato, Nancy M.},
title = {Using Guided Motion Planning to Study Binding Site Accessibility},
year = {2020},
isbn = {9781450379649},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3388440.3414707},
abstract = {Computational methods are commonly used to predict protein-ligand interactions. These methods typically search for regions with favorable energy that geometrically fit the ligand, and then rank them as potential binding sites. While this general strategy can provide good predictions in some cases, it does not do well when the binding site is not accessible to the ligand. In addition, recent research has shown that in some cases protein access tunnels play a major role in the activity and stability of the protein's binding interactions. Hence, to fully understand the binding behavior of such proteins, it is imperative to identify and study their access tunnels. In this work, we present a motion planning algorithm that scores protein binding site accessibility for a particular ligand. This method can be used to screen ligand candidates for a protein by eliminating those that cannot access the binding site. This method was tested on two case studies to analyze effects of modifying a protein's access tunnels to increase activity and/or stability as well as study how a ligand inhibitor blocks access to the protein binding site.},
booktitle = {Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics},
articleno = {109},
numpages = {10}
}
Abstract
Using Motion Planning to Evaluate Protein Binding Site Accessibility, Diane Uwacu, Everett Yang, Shawna Thomas, Nancy M. Amato, Technical Report, TR18-001, Parasol Laboratory, Department of Computer Science, Texas A&M University, College Station TX 77848, USA, Jul 2018. N/A Despite many efforts and considerable breakthroughs in ligand binding prediction, the best predictors still produce many false positives and a reliable,
fully automated prediction framework has yet to be developed. Binding site accessibility is an important feature ignored by methods that classify binding based solely on the energetic or geometric properties of the final bound protein-ligand complex. To evaluate this necessity, we transform the ligand accessibility problem into a robot motion planning problem where the ligand is modeled as a flexible agent whose task is to travel from outside the protein to its binding site. We use Rapidly-exploring Random Graphs coupled with Mean Curve workspace skeletons to quickly and thoroughly explore a protein environment in order to produce valid paths for ligand motion. Path weights reflect the influences of intermolecular forces on the given ligand. Low weight paths are extracted and analyzed for characteristics of accessibility. In this paper, we show that our algorithm provides a mechanism to evaluate binding site accessibility for a ligand.
Keywords: Ligand Binding, Motion Planning, Workspace Topology
Links : [Manuscript] BibTex
Abstract
